Comparative Genomics: I compared 123 cell wall associated proteins from Bacillus subtilis with proteins in 60 complete proteomes of a diverse collection of bacteria, including Gram positive and Gram negative species. Similar proteins found in many species of bacteria may be suitable targets for broad spectrum antibiotic drugs. There are at least two reasons for this: (1) proteins found in more species of bacteria are probably more important, and more likely to be essential, for bacterial survival than uncommom proteins; (2) if the normal function of such proteins can be inhibited by a chemical compound (drug), then that compound might be effective against a broad spectrum of bacteria. Broad spectrum antibiotics may be appropriate where the cause of an infection is unknown. For example, during an emergency where there is not enough time to make a specific diagnosis.
The cell wall is an esential structure for virtually all bacteria as it protects the cell from damage and osmotic lysis; it is the target of our best antibiotics (Leaver et al., 2009).
Given the importance of the cell wall to most bacteria and the lack of a cell wall in human cells, I focussed my research efforts on cell wall proteins. However, the cell wall is quite complex and there are over 100 cell wall associated proteins in B. subtilis. Starting with this well known model organism, I compared 123 B. subtilis cell wall protein sequences with proteins in 60 species of bacteria (selected for diversity and pathogenicity).
| Proteins with similar sequences tend to have similar 3D structures and similar functions in each organism (Rost, 1999). It is often said that sequence determines structure determines function, but small differences in sequence can make big differences in structure and, even more confusingly, very different sequences can fold into 3D structures that have the same function because they have the same active site (the place where other molecules bind to the protein)(Lesk, 2008). As a result, sequence analysis and predictions based on genomic data is often more about probability than certainty. Nonetheless, using BLAST (Altschul et al., 1997) for pairwise sequence alignment and the Protein Data Bank for 3D structure information, I generated the following and selected four proteins that are broadly conserved and have a known 3D structure. |
|
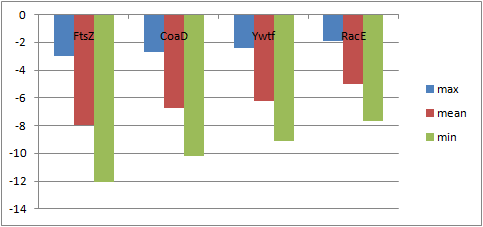
| Results: The docking scores for NPD ligands from the ZINC database bind best with FtsZ, with CoaD coming in second place. This chart shows the worst (max), the average (mean), and the best (min) binding score for each protein. These scores are in kcal/mol and, as odd as this may sound, the more negative the better. Based on the work completed so far, Demuris Ltd. has purchased the NPD chemical compounds with the greatest probability of success for in vitro testing. In theory at least, our research has provided a rational and principled basis for prioritizing relatively expensive wet lab experiments at virtually no cost per chemical compound (ligand) considered. |
 |